|
Gene Expression Core
Nalini Raghavachari, PhD,
Staff Scientist
Purpose and Contact Information
The purpose of the Gene Expression Core is to provide complete Genomics services to NHLBI investigators and the primary goals are to provide investigators with high quality, state-of-the-art gene expression profiling & genotyping services in a timely fashion using the Affymetrix platform by rigorous standardization of protocols and multiple quality control checks and to provide streamlined data analysis and identify signature genes by the application of complex statistical tools.
Services
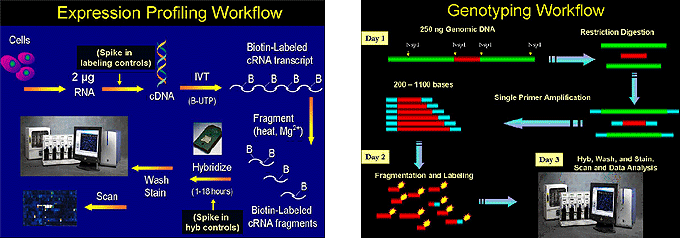
The Genomics Core Facility offers full service for expression profiling and genotyping using the Affymetrix platform. We will help you by recommending protocols for isolating RNA samples, assessment of RNA integrity and concentration, Real-Time PCR confirmation of results, workshops on data analysis for users, and most importantly one on one consultation during data analysis.
For more detailed information about the Gene Expression Core, please download this MS PowerPoint presentation: NR NHLBI GEP Core.ppt (Microsoft PowerPoint)
General Recommendations for RNA Preparation
It is critical to provide high quality mRNA or total RNA as the starting material for microarray analysis. We generally require atleast 2-3 µ g of total RNA at a concentration of 0.5 µ g per µ l. Problems we have found with user samples include:
- RNA degradation
- incorrect concentration
- contaminants (proteins, salt, genomic DNA, phenol, etc)
We recommend the following kits for total RNA isolation:
- RNeasy kits from Qiagen for total RNA ( > million cells)
- Ambion RNAqueous Micro Kit (<500, 000 cells)
If isolating the amount of recommended RNA is a problem, we recommend amplification of RNA using one of the commercially available kits such as Riboamp kit, Affymetrix kits.
In order for sample labeling and hybridization to work well, RNA must be of high quality. Contaminants such as phenol will inhibit reverse transcription reactions, consequently, we request that you DO NOT USE TRIZOL to extract your RNA unless you are confident you can remove all contaminants through subsequent chloroform extraction and ethanol precipitation. Submitted samples will be quality controlled using spectrophotometry and the Agilent bioanalyzer.
For more information on RNA isolation and sample submission please contact us.
The Affymetrix GeneChip System
Affymetrix GeneChips are commercially manufactured oligo-based arrays. Whereas traditional arrays are made by “printing” oligos or cDNA onto a microscope slide with capillary pins, the Affymetrix chips are made by directly synthesizing oligomers onto a silicon chip via photolithography. For more detail on the synthesis of the Affymetrix GeneChip, visit their web site at www.affymetrix.com.
Affymetrix GeneChips are synthesized by photolithography using 25-mer oligonucleotide probes. Each gene on the array is represented by 22 probes, 11 of which are perfect match (PM) and 11 of which are mismatch (MM). The mismatch probes are identical to the perfect match probes except for one nucleotide in the center of the oligo. The PM and the MM probe pairs allow quantitation and subtraction of signal caused by non-specific cross-hybridization. The GeneChips are designed so that the 11 probe pairs for each gene ( 11 PM and 11 MM) are randomly scattered about the GeneChip, which prevents an artifact from strongly affecting data for any one gene.
Real Time RT/PCR
Real Time RT/PCR is offered using the Applied Biosystems (ABI) Taqman system. ABI has designed primer/probe sets for most exons of human, mouse, and rat and offers them through the Gene Expression Assays program. The Genomics core facility will reverse transcribe and amplify the sample RNA and analyze the results using primer/probe sets purchased by the investigator from ABI.
RNA samples for Real-Time PCR analysis must be pure, RNA quality and purity is essential for the enzymatic reactions to work properly. Contaminants such as phenol will inhibit reverse transcription reactions; consequently, we request that you DO NOT USE TRIZOL to extract your RNA unless you are confident you can remove all contaminants through subsequent precipitation.
Notification of Results and Data Analysis
Users are notified by e-mail when QC data after hybridization is ready. Users who want to perform their own data analyses will be notified to collect their CD with raw data. Investigators can also be given access to Signet Server from where they can access their project data online.
The Core recognizes that many users will want help performing data analysis from multiple hybridizations (e.g. statistical significance, clustering, multi-dimensional scaling). Users wishing to meet individually for data analysis and consultation should contact Xiuli Xu to schedule an appointment once all the relevant hybridizations have been completed.
Contact Information
|