
Ken Kramer Ajay Thomas Kristin Castillo Chunhua Pan
|
|
Is the heparan sulfate fine structure a temporally and spatially permissive "heparan code"?
Heparan Sulfate (HS) is an unbranched sugar chain consisting of repeated disaccharides that can be modified at up to five positions, leading to an extraordinary level of sequence diversity. Commonly referred to as its fine structure, the pattern of HS modification over 2-7 disaccharides creates a specific ligand binding site. In many cases, a cell can only respond to a cell-cell signaling molecule if it has an appropriate HS fine structure at its cell surface. To characterize the HS fine structure, we are using directed evolution to bioengineer a set of peptides that bind to specific disaccharide units in the HS chain. One expectation is that we will be able to bring the peptides together in multiple combinations for specifically binding to larger HS saccharides. The hope is that this approach will help us define the 'HS code', a widely hypothesized model in which distinct HS modification patterns may provide information that is required for metazoan development.
|
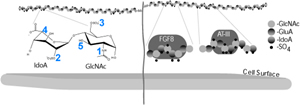
Figure: HS is a potentially information dense disaccharide repeat that can be modified at five positions (1 thru 5). Sulfation of uronic acid (IdoA/GluA) and glucosamine (GlcNAc) creates binding sites for a wide variety of extracellular proteins, fibroblast growth factor 8 (FGF8) and antithrombin III (ATIII) are illustrated as examples.
|