Automated
Genotyping
I.
Tissue Lysis
II. Generation
of Robot Source Files with FileMaker
III. Multiprobe
II HT PCR Set Up
IV.Caliper
AMS 90 SE Electrophoresis
V. Materials
and Recipes
I. Tissue Lysis:
- Pre-warm the Eppendorf Thermomixer R MTP to 55°C.
- Add 250 µL 20 mg/mL Proteinase K to a 10 mL thawed aliquot of
2mmLB.
- Spin down the plate (use a balance plate) in the Eppendorf 5804
centrifuge for 30 seconds at maximum possible rpm.
- Using a multi-channel pipetor and sterile trough containing the
2mmLB with Proteinase K. Transfer
100 µL of 2mmLB supplemented with Proteinase K to each well of a Nunc
96-well plate containing 2mm tail biopsies or other tissue samples to be
genotyped.
- Place the plate on the heating block for 1 minute uncovered and
without shaking to allow the lysis buffer to equilibrate to the proper
temperature.
- Seal the plate using a roller and new TemplateŇ
Sealing Foil.
- Replace the heating block cover and heat at 55°C with shaking at
1000 rpm for 4-6 hours. Check
the samples every hour to ensure the lysis is proceeding and that the
plate is still properly sealed.
An unsealed plated can result in lysis buffer evaporation. Do not lyse longer than 6 hours. If required, the Thermomixer R can be
programmed to change to 4°C without shaking for storage for a few hours
until the plate can be removed.
- Place the plate into the Thermocycler and run the Proteinase K
Inactivation Program before using lysates in PCR! After Proteinase K inactivation, the
plate can be stored frozen indefinitely until thawed for the next step
or keep it at 4°C or on ice for a few hours.
II.
Generation of Robot Source Files with FileMaker:
- Open the “Genotyping” FileMaker database consisting of the
following files: DNAList.fp5, MasterMix.fp5, Multiprobe.fp5,
PCRList.fp5, and PostMix.fp5. The
DNAList.fp5 file will be brought to the foreground.
- Import or enter into the DNAList.fp5 file the listing of DNA
sample records on 96-well plates with the following parameters included
for each sample: TagID, 96-well plate date ID, well column, and well row. Each sample should also have each
Genotype TBD listed as needed (up to three). This list can be imported from a comma or tab delimited
text file. The database will
enter the PCR reactions required for each Genotype TBD by looking up the
PCR values specified for each GenotypeTBD from the PCRList.fp5
file. The database will also
calculate the Multiprobe well number for each sample from the column and
row information. Creating new
records and entering the required info on the “Add Samples” layout can
also add DNA sample records.
- Run the “Find DNA Plate Records” script to find all records that
are to be processed. Up to four
plates of DNA can be processed at once on the Multiprobe II HT.
- Run the script: “Generate Multiprobe Table”. This script will create a file of the
reactions required. In English,
the script performs the following:
- Assigns a DNA Plate# to each 96-well
plate containing DNA samples.
- Deletes
previous sample records from the “Multiprobe.fp5” file.
- Goes
through each found record’s PCR fields (1A-3B). If there is a reaction specified, a
record is added to the “Multiprobe.fp5” file for each instance with DNA
well number, PCR reaction required, DNA TagID, DNA Plate#, and DNA
Plate ID recorded for each PCR reaction.
- Finds,
sorts, and numbers the needed PCR reactions required plus ‘H2O’
in the “MasterMix.fp5” file.
- Adds
(-) controls for each PCR reaction required.
- Adds
‘H2O’ wells to fill up the last used row of the PCR plate
(required for Caliper AMS 90 SE).
- Checks
that the number of samples requires no more than four PCR plates.
- Sorts
by PCR required and TagID (ascending)
- Assigns
PCR plate # (up to 4), well row, and well column numbers arranged for
Caliper AMS 90 SE processing in rows.
The
resulting “Multibrobe.fp5” file is a set of records for each PCR reaction
required for all the found DNA samples.
In addition to the field values entered by the script, the file also
includes a number of additional fields whose values are calculated based on
the information recorded for each record (well numbers, field value counts,
and MasterMix.fp5 lookups) or are constant for most records.
- Run the script: “Print Master Mix List”. This will bring up a dialog box for
 printing a listing of all
the PCR master mixes required for the current set of DNA samples with
the correct volumes calculated based on a count of each reaction and the
recipe information entered in “MasterMix.fp5”. printing a listing of all
the PCR master mixes required for the current set of DNA samples with
the correct volumes calculated based on a count of each reaction and the
recipe information entered in “MasterMix.fp5”.
- Run the script: “Export Multiprobe Source Data”. Follow the directions in the message
to export a listing of PCR reaction records sorted by MasterMixVolume,
(descending), PCRPlate (ascending), PCRMultiprobeWell (ascending), and
DNASource (descending) as comma-separated values that the Multiprobe II
software will access to set up the PCR reactions.
- Run the script: “Export Caliper Source Data”. Follow the directions in the message
to export a listing of PCR reaction records sorted by PCRPlate
(ascending) and PCRCaliperWell (ascending) as comma-separated values
that can be imported into the Caliper AMS 90 SE software as sample information. If you are setting up more than one
PCR plate you will have to run this script once for each plate.
III.
Multiprobe II HT PCR Set Up:
 Prepare
the PCR Master Mixes as listed on the printout from step 5 of the
previous section. Prepare
the PCR Master Mixes as listed on the printout from step 5 of the
previous section. - Spin down the DNA plate(s) (use a balance plate) in the
Eppendorf 5804 centrifuge for 5 minutes at maximum possible rpm.
- Start the Multiprobe Winprep II software.
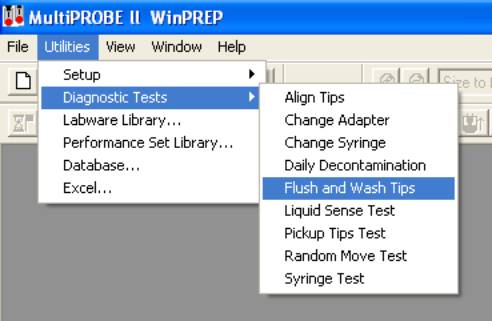
- Initialize the instrument.
- Run Flush and Wash Tips utility until there are no more air
bubbles visible in the Multiprobe II HT’s tubing.
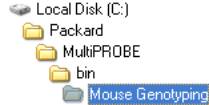 Open
the “MousePCR.MPT” file located in the Mouse Genotyping Folder in the
path shown on the right. Open
the “MousePCR.MPT” file located in the Mouse Genotyping Folder in the
path shown on the right.- Place the Nunc DNA plate(s) on the deck of the Multiprobe II HT
at the labware positions corresponding to DNA1, DNA2, DNA3, DNA4 as
listed in the “PlateID List” layout of the “Multiprobe.fp5” file.
- Place new MJ Research Microseal 96-well plates on the deck of
the Multiprobe II HT at the labware positions corresponding to PCR1,
PCR2, PCR3, up to PCR4 as needed for the number of reactions to be set
up.
 Load
the master mix tubes onto the 1.5 mL adapter plate as listed in the
master mix printout. The well
map appears on the right. Load
the master mix tubes onto the 1.5 mL adapter plate as listed in the
master mix printout. The well
map appears on the right.- Load tip boxes onto the back row of the deck as listed (two
boxes of 200 µL tips on the left and two boxes of 20 µL on the right).
- Start the run by clicking on “Execute”.
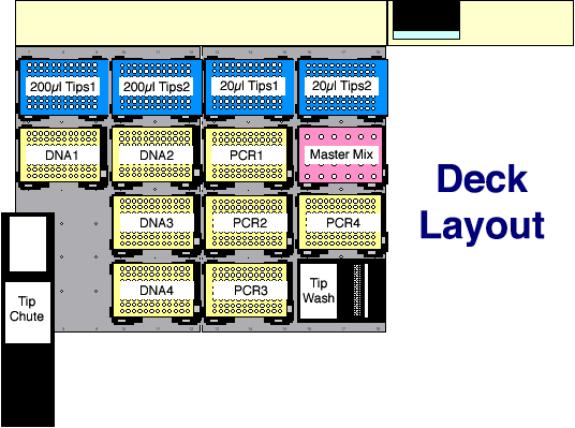
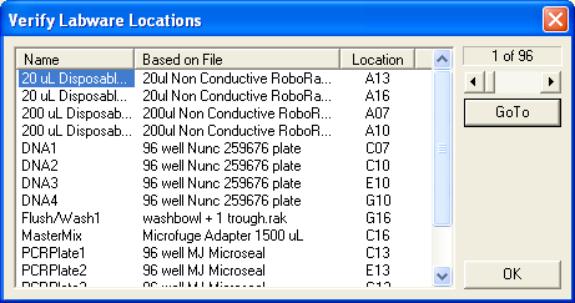
- An “Initial User Query” box will be displayed. Click on “Verify
Labware Locations” to bring up the “Verify Labware Locations” box.
- Click on each labware name in turn and click on “GoTo”. The robot arm should move to the
specified labware item and lower multiprobe tip #1 to the first well of
the labware’s well map. Check to
make sure that the tip lines up in the center of the first well
location. If it doesn’t, it will
be necessary to reevaluate the labware (see Multiprobe II HT
documentation for details on how to define and evaluate labware). When all labware that will be used
has been checked, click on “OK”.
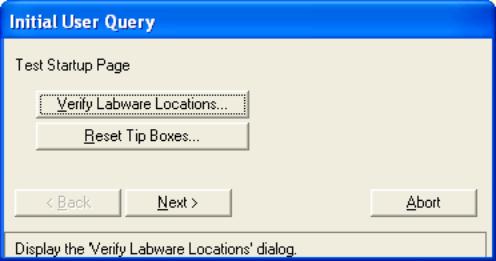
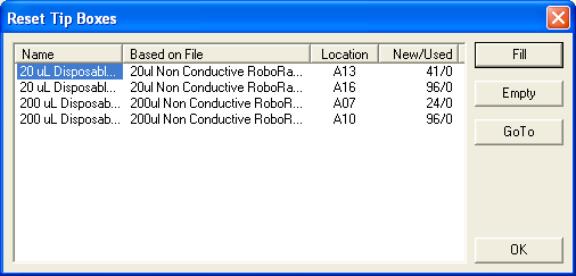
- Back at the “Initial User Query” box click on “Reset Tip Boxes…”
to display a box for resetting tips.
The numbers should match up to what is actually present in each
tip box. If they do not, select
the offending tip box’s name and click on “Fill”. This will generate an error in
finding tips later, during the run.
Then, during the run when the error occurs, it will be necessary
to repeatedly select “Another Tip” from the “Tip Pickup Error” box until
the robot is able to pickup up a full set of unused tips. Click on “OK” when you are done
resetting tips.
- Back at the “Initial User Query” Click on “Next”.
- A new “Initial User Query” box will appear that states “please
browse for file”.
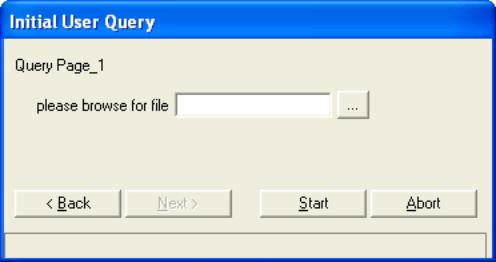 Click on “…”. Locate the .csv file of the PCR
created in step 6 of the “Generate Robot Source Files” section of this
document. Click on “…”. Locate the .csv file of the PCR
created in step 6 of the “Generate Robot Source Files” section of this
document.
- When you have selected the correct file, click on “Start”, and
the Multiprobe II HT will start the pipetting steps.
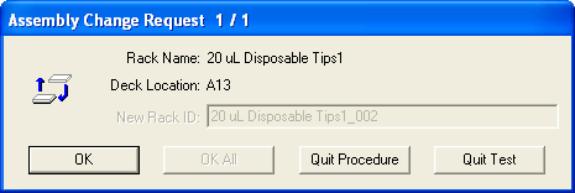
- If you are setting up more PCR wells than there are tips present
on the rack, a prompt will show up when the required tips are used
up. Change the specified deck
location’s box with new tips and click OK.
- Once all the PCR wells are prepared, top each PCR plate with
12-strip caps. Place in a
96-well plate alpha unit on an MJ Research Thermocycler and run Program
58.
IV. Caliper
AMS 90 SE Electrophoresis
See HT DNA 5000 SE 30 Reagent
Kit Insert and Section V Required Materials and Recipes!
V. Materials
and Recipes:
- 200 µL Nunc 96-well plates (#259676) for tail biopsy lysis
- USA Scientific TemplateŇ
Sealing Foil (#2923-0100) for sealing Nunc plates.
- MJ Research Microseal 96-well plate (MSP-9601) for PCR.
- MJ Research 12-Strip Caps (TCS-1201) for sealing Microseal
plates
- 2mmLB (Nagy, A. 2003. Manipulating the Mouse Embryo: A
Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring
Harbor, NY).
|
|
50
|
mM KCl
|
25
|
mL
|
2
|
M KCl
|
|
|
1.5
|
mM MgCl2
|
0.75
|
mL
|
2
|
M MgCl2
|
|
|
10
|
mM Tris
|
10
|
mL
|
1
|
M Tris-HCl pH8.3
|
|
|
10 mg
|
Gelatin
|
1
|
mL
|
10
|
mg/mL Gelatin
|
|
|
0.045%
|
NP40
|
.45
|
mL
|
100
|
% NP-40
|
|
|
0.045%
|
Tween-20
|
.45
|
mL
|
100
|
% Tween-20
|
|
|
|
H2O
|
945.25
|
mL
|
|
H2O
|
|
|
|
|
1000
|
mL
|
|
|
Autoclave in 4 X 250 mL bottles.
Store unopened bottles at room temperature. Otherwise keep 10 mL single use aliquots per 15 mL tube at
–20°C. 10 mL is enough for lysis of a
single full 96-well plate.
- 20 mg/mL Proteinase K
- Multiprobe II HT clear disposable 200 µl and 20 µl tips (one of
each for each PCR well).
- Proteinase K Inactivation program conditions:
|
1
|
95°C
|
10:00 minutes
|
|
2
|
4°C
|
forever
|
|
3
|
End
|
|
- Thermocycling program 58 conditions:
|
1
|
95°C
|
5:00 minutes
|
|
2
|
95°C
|
0:45 seconds
|
|
3
|
58°C
|
0:30 seconds
|
|
4
|
72°C
|
0.4°/second
|
|
5
|
72°C
|
0:30 seconds
|
|
6
|
Goto 2
|
39 times
|
|
7
|
72°C
|
10:00 minutes
|
|
8
|
4°C
|
forever
|
|
9
|
End
|
|
- Caliper AMS90 HT DNA 5000 SE 30 chip
- Caliper AMS90 HT DNA 5000 SE 30 reagents (#760124).
- 1X PCR Buffer Solution for Buffer Trough:
|
|
4
|
mL
|
25 mM MgCl2
|
|
|
5
|
mL
|
10X PCR Buffer
|
|
|
41
|
mL
|
H2O
|
|
|
50
|
mL
|
|
- HT DNA 5000 Ladder for Ladder Trough well “A”:
|
|
12
|
µL
|
HT DNA 5000
|
|
|
12
|
µL
|
10X PCR Buffer
|
|
|
9.6
|
µL
|
25 mM MgCl2
|
|
|
86.4
|
µL
|
H2O
|
|
|
120
|
µL
|
|
1. Transfer
1 mL of HT DNA 5000 SE 30 Gel Matrix (red cap) and 25 µL of HT DNA 5000 SE
Dye Concentrate (blue cap) to a 1.5 mL microcentrifuge tube.
2. Vortex
the solution until it is well mixed.
Transfer the mixture to two spin filters (500 µL each).
3. Centrifuge
at RCF = 900 X G at room temperature for 10 minutes or until Gel-Dye Mix has
passed through the filter. Record the
date on the tube. Store in the dark
at 4°C. Use within 3 weeks.
- Caliper Chip Storage Buffer (0.2µm filtered):
|
|
200 mM TAPS
|
|
|
2 mM EDTA pH 8.0
|
|